New method speeds up and improves accuracy of medical-image atlas production
by
John R. Fischer, Senior Reporter | December 13, 2019
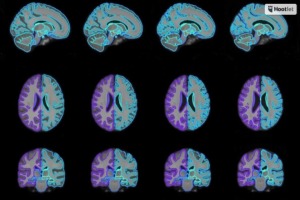
Generating more accurate atlases for medical image analysis in diagnostics may be possible with the development of a new method by researchers at the Massachusetts Institute of Technology (MIT).
The technique utilizes machine learning to speed up the often lengthy process of atlas production to create a more customized "conditional" atlas that takes into account specific patient attributes, such as age, sex and disease. Customization is based on shared information from across the entire data set used, enabling the model to synthesize atlases from patient subpopulations that may be completely missing in a data set.
“The world needs more atlases,” says first author Adrian Dalca, a former postdoc in the Computer Science and Artificial Intelligence Laboratory (CSAIL) and now a faculty member in radiology at Harvard Medical School and Massachusetts General Hospital. “Atlases are central to many medical image analyses. This method can build a lot more of them and build conditional ones as well.”
Using medical image data sets to construct structural relationships that show disease progression, atlas-building is an iterative optimization process that can take days or even weeks to complete. Aligning scans to an initial and often blurry atlas, the process computes a new average image from the aligned scan and is then repeated for all images until a final atlas is produced. As a result, many researchers download and use publicly available atlases to speed up the work, but at the cost of capturing distinct details of individual data sets or specific subpopulations.
Dalca and his colleagues sought to address this by combining two neural networks into one that rapidly aligns images and speeds up the process, with one network automatically learning an atlas at each iteration and the other simultaneously aligning that atlas to images in a data set.
Training consisted of feeding the joint network a random image from a dataset encoded with desired patient attributes. The joint network then estimated an attribute-conditional atlas, with the second network aligning this estimate with the input image and generating a deformation field, which is used to capture structural variations.
The deformation field can train what is called a "loss function", a component of machine learning models that helps minimize deviations from a given value. The function can then be used to interpret how specific attributes correlate to structural variations across all images in a data set. When all attributes are taken into account, the network is able to use all learned information across the data set to synthesize an on-demand atlas, even if attribute data is missing or lacking within the dataset.
The team hopes that clinicians can use the method to build their own atlases fast from their own and potentially small data sets. The aim, according to Dalca, who is working with MGH researchers to harness a data set of pediatric scans to generate conditional atlases for children, is to eventually have one function able to produce conditional atlases for any subpopulation, from birth to 90 years old. Researchers will simply log into a web page, and input age, sex, diseases, and other parameters to produce an on-demand conditional atlas.
“That would be wonderful, because everyone can refer to this one function as a single universal atlas reference,” he said.
The team verified the function developed in its research by generating conditional templates for various age groups between 15 and 90. It will present its findings in December at the Conference on Neural Information Processing Systems.
|
|
|
You Must Be Logged In To Post A Comment
|
